Analysis of Genetic Structure in the USDA Rice Mini-Core Collection Using the SQUAMOSA Promoter-binding-like Gene Family 
2. Sichuan Normal University, Chengdu, 610066, P.R. China
3. Dale Bumpers National Rice Research Center, United States Department of Agriculture–Agricultural Research Service(USDA-ARS), 2890 Highway 130 East, Stuttgart, AR 72160, USA
* The authors contributed equally to the work
 Author
Author  Correspondence author
Correspondence author
Rice Genomics and Genetics, 2012, Vol. 3, No. 1 doi: 10.5376/rgg.2012.03.0001
Received: 16 Dec., 2011 Accepted: 17 Jan., 2012 Published: 20 Jan., 2012
Ren et al., 2012, Analysis of Genetic Structure in the USDA Rice Mini-Core Collection Using the SQUAMOSA Promoter-binding-like Gene Family, Rice Genomics and Genetics, Vol.3, No.1, 1-7 (doi: 10.5376/rgg.2012.03.0001)
The SQUAMOSA (SQUA) promoter-binding-like (SPL) gene family is a plant specific transcription factor family and has many important biological functions. In this study, 19 pairs of primers were designed based on 19 Oryza sativa L. SQUAMOSA promoter-binding-like (OsSPL) gene sequences and used for a polymorphism analysis with 171 accessions as a set of the United States Department of Agriculture (USDA) rice mini-core collection established in the US. This mini-core represents the core of 1 794 accessions and the entire collection of 18 412 accessions. In total, 140 bands were amplified, and 128 of them were polymorphic, occupying a great majority (up to 91.4%) of the mini-core collection. The average number of alleles per locus was 7.4. The cluster and principal coordinate analysis agreeably classified the 171 accessions into six subgroups (subgroups â… -â…¥), and those in the same subgroup were distributed throughout similar geographical and ecological regions worldwide. Days to heading for subgroup â…¥ was significantly greater than the overall mean. Similarly, the plant height for subgroup â…¡ and spikelets per panicle for subgroups â… and â…¢ were significantly different from the overall mean, respectively. Thus, the OsSPL genes were associated with the days to heading, plant height and spikelets per panicle.
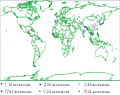
Rice is a staple food for nearly half of the world's population, and there are more than 100 000 rice varieties planted worldwide that provide almost one-quarter of the global per capita energy (Sasaki, 2008). The main agronomic traits in rice, such as the grain number per spike, 1 000-grain weight and tillering ability, directly influence rice yield (Gao et al., 2007; Wang et al., 2009; Yang et al., 2000; Miura et al., 2010). The SQUAMOSA promoter binding protein-like (SPL) gene family contains a highly conservative DNA domain that is representative of families of plant-specific transcription factors and participates in the adjustment of flowering-time, the transition of flower development, the initiation of leaf development and other developmental processes (Birkenbihl et al., 2005; Wang et al., 2008; Wang et al., 2009; Dai et al., 2010; Miura et al., 2010; Jiao et al., 2010). The Oryza sativa L. SQUAMOSA promoter binding protein-like 14 (OsSPL14) gene has been cloned and proved to involve in controlling the tiller number in the vegetative developmental stage, whereas elevated expression of OsSPL14 in the reproductive developmental stage will promote branching, which resulted in a reduction of tiller number, an increased number of spikelets and grains per panicle, a thickened haulm, increased lodging resistance, and enhanced grain yields (Jiao et al., 2010; Miura et al., 2010). There are 19 OsSPL genes annotated in the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). And all 19 genes expression has been confirmed in rice (Xie et al., 2006). In this study, 19 pairs of OsSPL gene primers were designed according to their sequences and used for a polymorphism analysis of 171 accessions (Agrama et al., 2009) in a United States Department of Agriculture (USDA) rice mini-core collection established in the US. We analyzed the gene-trait association between the genotypes and phenotypes to elucidate the association between OsSPL gene polymorphisms and the major agronomic traits. Figure 1 Global distribution of 171 accessions and six subgroups of the USDA rice mini-core collection
1 Results and Analysis
1.1 Global distribution of the 171 accessions and their composition
The 171 accessions in the USDA rice mini-core collection consist of 163 Asian cultivars (Oryza sativa), 7 African cultivars (Oryza glaberrima), and 1 common wild rice (Oryza rufipogon griff.); these cultivars are distributed throughout 68 countries (Figure 1). There are 93 accessions in Asia (~54.4%), with 19 China's accessions, the most among all of the countries, and 13 India's accessions (the second most). Other countries in globally have 1~3 accessions.
1.2 Sequence analysis between the Japonica and Indicia OsSPL genes
Single nucleotide mutations and/or cantlet-base deletions were identified between the Keng (Japonica) Nipponbare OsSPL genes and Hsien (Indica) 9311 OsSPL DNA sequences based on http://www.gramene.org/Multi/blastview/ BLA_K8FkdkRLV. A pairwise protein alignment showed that the identity in protein sequences of the Keng (Japonica) Nipponbare OsSPL genes and Hsien (Indica) 9311 OsSPL genes was 100%, which indicated no difference between the coding sequences of the Keng (Japonica) Nipponbare and Hsien (Indica) 9311 OsSPL genes.
1.3 PCR assay of 171 accessions
In this study, 19 pairs of OsSPL gene primers were designed, and 140 bands were amplified totally. Among them, 128 were polymorphic, comprising a majority (up to 91.4%) of the mini-core collection. The average number of alleles per locus was 7.4 (Table 1).
Table 1 Primers for the OsSPL genes and the polymorphic loci for genotyping of 171 accessions in the USDA rice mini-core collection |
1.4 Cluster and principle coordinate analysis
Using the unweighted pair group method with arithmetic mean (UPGMA), our cluster analysis based on the PCR results agreeably classified the 171 accessions into two groups when using a simple matching coefficient of 0.65. The first group consisted of 10 accessions, and the second group consisted of 161 accessions. At the simple matching coefficient of 0.687, the first group comprised one subgroup â… , and the second group was divided into five subgroups (Figure 2). Subgroups â…¡-â…¥ consisted of 20, 48, 45, 24, and 24 accessions, respectively. The 7 African cultivars were classified into three subgroups: USDA IDs 321 and 1688 belonged to subgroup â…¢, USDA IDs 1689, 1691, 1693, and 1694 were categorized into subgroup â…¤, and USDA ID 1692 was put in subgroup â…£. Common wild rice (USDA ID 1703) was classified into the â…¥ subgroup. Thus, there was no obvious differentiation between the Asian and African cultivars in the OsSPL gene family.
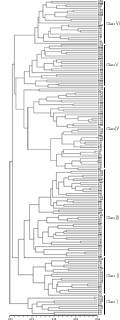 Figure 2 Dendrogram of 171 accessions in the United States Department of Agriculture (USDA) rice mini-core collection generated by cluster analysis of genetic distance from 19 OsSPL primers |
The results of the principal coordinates analysis based on the original matrix of the PCR amplification indicated that the first two and first three coordinates could explain 70.21% and 72.17% of their correlation, respectively. Two-dimensional and three-dimensional scatter plots of the 171 accessions were generated (Figure 3; Figure 4), showing a strip-type discrete distribution in the two-dimensional diagram that was clearly divided into two groups and six subgroups; these groupings were more clearly demonstrated in the three-dimensional diagram. The three classification results were fundamentally consistent; moreover, the three-dimensional diagram based on the principal coordinate analysis showed the relationship among the 171 accessions at different directions and levels.
 Figure 3 Scatter plots of the first and second principal coordinates of the USDA rice mini-core 171 accessions generated from the principal coordinates analysis on polymorphisms by 19 OsSPL primers |
 Figure 4 Three dimensional plots of the first and second principal coordinates of the USDA rice mini-core 171 accessions generated from the principal coordinates analysis on polymorphisms by OsSPL primers |
1.5 Global geographical distribution of the subgroups the 171 accessions
The geographical distribution of the subgroups of the 171 accessions indicated that the accessions of each subgroup showed strongly similar geographical and ecological distributions and were not randomly distributed worldwide. For example, the accessions of subgroup â… were distributed in tropical rice-growing areas, with the exception of several accessions that were distributed throughout China. Accessions of the same subgroup showed a high frequency in similar geographical rice-growing areas, such as those in Panama, Costa Rica, and Honduras, belonging to the Central American area of subgroup â…£.
1.6 Gene-trait association analysis
Based on the results of the cluster analysis, the mean of the phenotypic data of each subgroup was calculated. A single sample mean test was analyzed between the mean of each subgroup and the mean of the phenotypic data of the total sample, which consisted of the 171 accessions, namely, the gene-trait association.
The gene-trait association (Table 2) showed that there was a significant difference between the mean of the spikelets per panicle of subgroup â… and the mean of the spikelets per panicle of the total sample as well as a significant difference between the mean of the plant height of subgroup â…¡ and that of the total sample. There was also a significant difference between the mean of the spikelets per panicle of subgroup â…¢ and that of the sample as well as a significant difference between the mean of the days to heading of subgroup â…¥ and that of the sample. The mean of the plant height of subgroup â…¤ and that of the sample almost reached a significant level, with a difference probability of 0.052 (P=0.052). There was no significant difference between the mean of each trait of subgroup â…£ and that of the sample, which indicated that the mean of each trait of subgroup â…£ was no offset. There was no significant difference in the mean of the tillers per plant or brown 1 000-grain weight between each subgroup and the total sample, which indicated that the traits of tillers per plant and the brown 1 000-grain weight of each accession of the subgroups were random. As shown in Table 2, the standard deviation of each trait of each subgroup was essentially similar to the standard deviation of each trait of the total sample except that for the days to heading and spikelets per panicle of subgroup â…¤, which indicated that the classification was reliable. Thus, according to the Hardy-Weinberg equilibrium, the OsSPL genes were associated with the traits of spikelets per panicle, plant height and days to heading.
Table 2 Single sample mean value examination on phenotype data mean of subgroup |
2 Discussion
2.1 Polymorphism of the OsSPL gene family
As a plant-specific transcription factor family, the SPL gene family regulates many important biological functions (Birkenbihl et al., 2005; Wang et al., 2008; Wang et al., 2009; Dai et al., 2010; Miura et al., 2010; Jiao et al., 2010). Rice is a model plant in genomics research, and the full genome sequence has been drafted. At present, rice functional genomics shifts to focus on gene function research and gene diversity analysis. The USDA Rice Core Collection consists of 1 794 accessions, which represent 18 412 rice germplasm samples from 116 countries, essentially covering the global rice-growing regions. 88% reliability that the information in the USDA Rice Core Collection is representative of the entire rice germplasm has been reported (Yan et al., 2007). In the present study, a subset of the Core Collection, 171 accessions comprising a rice mini-core collection, suggested genetic diversity among the rice materials. OsSPL primers were designed according to 19 OsSPL gene fragment sequences to surely amplify corresponding OsSPL target genes, and the PCR results indicated that the sequences showed higher rates of polymorphism, with the exception of primers SPL 15 and SPL 19. Xie et al (2006) reported that the OsSPL gene expression analysis using real-time RT-qPCR indicated that the OsSPL1-OsSPL18 genes were classified into three groups. The first group was highly expressed in the spike, the second group was expressed in all organs and highly expressed in the leaf, leaf sheath and panicle, and the third demonstrated no differential expression in different organs. And it has been reported that 11 of the OsSPL genes were regulated by miRNA 156. In the present study, we found little variation in the amplification of the OsSPL genes with expressing uniformly in all organs and/or little regulation by miRNA 156, and thus, suggesting little variation in evolutionary processes between the accessions.
Based on the results of the PCR amplification using the OsSPL primers, the 171 accessions were classified into two groups and six subgroups. Furthermore, the accessions of each subgroup showed a similar eco-geographical distribution, which suggested that their distribution had some connection with long-term human selection and ecological adaptation for cultivation. We found no difference between the Asian and African cultivars. When comparing the Keng (Japonica) Nipponbare OsSPL genes and Hsien (Indica) 9311 OsSPL genes, there were single base mutations or cantlet-base deletions in the DNA sequence, though no differences in the protein sequence, which might be due to the OsSPL genes being indispensable transcription factors for rice growth and development. Their functions have been reported only to be regulated post-transcriptionally (Xie et al., 2006; Dai et al., 2010). We did not determine whether the similar eco-geographical distribution had any connection with the differentiation of Hsien (Indica) and Keng (Japonica) rice because we did not obtain detailed classification information on the Asian cultivars. Further studies are required to address this issue.
2.2 Gene-trait association
According to the Hardy-Weinberg equilibrium theory, the OsSPL genes were associated with the traits of spikelets per panicle, plant height, and days to heading; these results were obtained by testing the mean of each subgroup using a single sample mean test and were essentially consistent with previous studies (Miura et al., 2010; Jiao et al., 2010). In this study, it was not found that the OsSPL genes controlled tillering and the brown 1 000-grain weight. We speculate that the major cause of inconsistency might be that tillering is easily influenced by the environment, whereas the brown 1 000-grain weight is less affected by the environment. Thus, further research is required to investigate the role of the OsSPL genes in controlling the brown 1 000-grain weight.
3 Materials and Methods
3.1 Test materials and data acquisition
Rice materials used in this study were provided by our collaborator from Dale Bumpers National Rice Research Center, Agricultural Research Service (ARS) of the United States Department of Agriculture (USDA), and these materials had been studied preciously (Yan et al., 2007; Agrama et al., 2009). In April 2010, the materials were planted at the Xindu Testing Site of Sichuan Academy of Agricultural Sciences. The experiment was set up three replicates that three rows in each replicate and seven plants in each row, with the planting and rowing space of 16 cm×26 cm. In the early June 2010, 5 plants in the midst of the midst row in each replicate were sampled for their tillers. The remaining data was collected from the data in 2006 (Yan et al., 2007).
3.2 Primers design and PCR amplification
The OsSPL primers were designed according to 19 sequences of OsSPL genes downloaded from http://rice.plantbiology.msu.edu/ using Primer Premier 6.0 (Table 1).
The CTAB method was used to extract DNA from the rice leaves using (Murray and Thompson, 1980) with slight modifications. PCR amplification was performed according to a previously described method in our lab (Gao et al., 2008), and the annealing temperatures of the primers were listed in Table 1.
3.3 Data processing
The means of phenotypic data of each accession were calculated, and significance tests were performed using DPS 7.0 software. The PCR amplification bands were recorded by referring to the presence of a band as “1” and the absence of a band as “0”. The data analyses were performed using NTSYpc-2.0e software.
According to the Hardy-Weinberg equilibrium, the gene frequency of a sample randomly selected from a random population indicates no significant difference. In other words, the mean of the phenotypic data of the sample is not significantly different from that of the population. Based on the results of cluster analysis using PCR amplification, the phenotypic data mean of each subgroup and the phenotypic data mean of the 171 accessions were calculated to test whether there was any significant difference between them. Then these results were used to judge if any traits were linked to the OsSPL genes, namely, the gene-trait association analyses.
3.4 OsSPL genes DNA and amino acid sequences alignments
A DNA sequence alignment was performed between the Keng (Japonica) OsSPL genes and Hsien (Indica) OsSPL genes using http://www.gramene.org/Multi/blastview/BLA_K8FkdkRLV. The amino acid alignment was carried out using the amino acid sequences of all of the OsSPL genes downloaded from http://pfam.janelia.org/family/PF03110.7#tabview= tab2 using DNAMAN 6.0 software.
Authors' contributions
Guangjun Ren and Juansheng Ren conceived of the project and its components. Juansheng Ren, Yuchao Yu and Guangjun Ren contributed to the original concept of the project. Fangyuan Gao, Lihua Zeng, Xianjun Lu, Xianting Wu and Wengui Yan collected samples, performed the phenotyping and corrected the paper. Juansheng Ren, Yuchao Yu and Guangjun Ren analyzed all of the data together and wrote the paper. All authors have read and approved the final manuscript.
Acknowledgments
This research project was supported by grants from the “948” project of Ministry of Agriculture of the People's Republic of China (2006-G1), National Natural Science Foundation of China (30900891), China Agricultural Research System and National High-tech R&D Program of China (863 Program). We also thanked two anonymous peer reviewers for their assessments and advice.
Reference
Agrama H.A., Yan W.G., Lee F., Fjellstrom R., Chen M.H., Jia M., and McClung A., 2009, Genetic assessment of a mini-core subset developed from the USDA Rice Genbank, Crop Science, 49: 1336-1346
http://dx.doi.org/10.2135/cropsci2008.06.0551http://dx.doi.org/10.2135/cropsci2008.06.0551er
Birkenbihl R.P., Jach G., Saedler H., and Huijser P., 2005, Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains, J. Mol. Biol., 352(3): 585-596
http://dx.doi.org/10.1016/j.jmb.2005.07.013 PMid:16095614
Dai F.G., Hu Z.L., Chen G.P., Wang B.Q., and Wang Y., 2010, Progress in the plant specific SBP-box gene family, Chinese Bulletin of Life Sciences, 22(2): 155-160
Gao F.Y., Ren G.J., Lu X.J., Sun S.X., Li H.J., Gao Y.M., Luo H., Yan W.G., and Zhang Y.Z., 2008, QTL analysis for the resistance to pre-harvest sprouting in rice (Oryza sativa L.), Plant Breeding, 127(3): 268-273
http://dx.doi.org/10.1111/j.1439-0523.2007.01450.x
Gao L.Y., and Zhou H.F., 2007, Relationship between yield component factors and yield in rice, Liaoning Agricultural Sciences, 1: 26-28
Jiao Y.Q., WangY.H., Xue D., Wang J., Yan M.X., Liu G.F., Dong G.J., Zeng D.L., Lu Z.F., Zhu X.Z., Qian Q., and Li J.Y., 2010, Regulation of OsSPL14 by osmir156 defines ideal plant architecture in rice, Nature Genetics, 42(6): 541-544
http://dx.doi.org/10.1038/ng.591 PMid:20495565
Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., and Ashikari M., 2010, OsSPL14 promotes panicle branching and higher grain productivity in rice, Nature Genetics, 42(6): 545-549
http://dx.doi.org/10.1038/ng.592 PMid:20495564
Murray R.A., and Thompson W.F., 1980, Rapid isolation of high molecular weight plant DNA, Nucleic Acids. Res., 8(19): 4321-4325
http://dx.doi.org/10.1093/nar/8.19.4321 PMid:7433111 PMCid:324241
Sasaki T., 2008, From the editor’s desk, Rice, 1(1): 1-2
http://dx.doi.org/10.1007/s12284-008-9010-y
Wang J.W., Czech B., and Weigel D., 2009, Mir156-regulated spl transcription factors define an endogenous flowering pathway in Arabidopsis thaliana, Cell, 138(4): 738-749
http://dx.doi.org/10.1016/j.cell.2009.06.014 PMid:19703399
Wang J.W., Schwab R., Czech B., Mica E., and Weigel D., 2008, Dual effects of mir156-Targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana, The Plant Cell, 20(5): 1231-1243
http://dx.doi.org/10.1105/tpc.108.058180 PMid:18492871 PMCid:2438454
Wang Z.G., Wang L., and Liao X.Y., 2009, Characteristic analysis on yield components of the medium Indica rice from regional trials in China, Acta Agriculturae Zhejiangensis, 21(4): 345-349
Xie K.B., Wu C.Q., and Xiong L.Z., 2006, Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice, Plant Physiology, 142(1): 280-293
http://dx.doi.org/10.1104/pp.106.084475 PMid:16861571 PMCid:1557610
Yan W.G., Rutger J.N., Bryant R.J., Bockelman H.E., Fjellstrom R.G., Chen M.H., Tai T.H., and McClung A.M., 2007, Development and evaluation of a core subset of the USDA Rice Germplasm Collection, Crop Science, 47(2): 869-878
http://dx.doi.org/10.2135/cropsci2006.07.0444
Yang R.F., Xia Z.Y., and Zhang C.H., 2000, Genetic cluster of plant-type trails of rice varieties, Journal of Anhui Agrotechnical Teachers College, 14(1): 61-64
. PDF(934KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Juansheng Ren
. Yuchao Yu
. Fangyuan Gao
. Lihua Zeng
. Xianjun Lu
. Xianting Wu
. Wengui Yan
. Guangjun Ren
Related articles
. Rice
. SQUAMOSA promoter-binding-like ( SPL ) genes family
. Germplasm
. Core collection
. Gene-trait association
Tools
. Email to a friend
. Post a comment


